RESEARCH ARTICLE
Biofabricated Silver Nanoparticles Synergistically Activate Amphotericin B Against Mature Biofilm Forms of Candida Albicans
Shivkrupa D. Halbandge, Supriya P. Mortale, Sankunny Mohan Karuppayil*
Article Information
Identifiers and Pagination:
Year: 2017Volume: 4
First Page: 1
Last Page: 16
Publisher Id: TONMJ-4-1
DOI: 10.2174/1875933501704010001
Article History:
Received Date: 27/04/2017Revision Received Date: 27/07/2017
Acceptance Date: 10/08/2017
Electronic publication date: 30/09/2017
Collection year: 2017
open-access license: This is an open access article distributed under the terms of the Creative Commons Attribution 4.0 International Public License (CC-BY 4.0), a copy of which is available at: https://creativecommons.org/licenses/by/4.0/legalcode. This license permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Background:
Biofilm formation by Candida albicans is a significant clinical challenge. Fungal biofilms are resistant to most of the currently available antifungal agents. Amphotericin-B (AmB) is an antifungal agent used for the treatment of systematic fungal infections but it is well known for its toxicities and side-effects. Novel approaches are needed to treat these infections that can reduce its toxicities.
Objectives:
Current study aims to evaluate the efficacy of silver nanoparticles (SNPs) alone and in combination with AmB against growth and biofilm formation in C. albicans.
Methods:
Combinations of SNP-AmB were tested against planktonic growth and biofilm formation in vitro. Micro broth dilution method was used to study planktonic growth and biofilm formation. The fractional inhibitory concentration indices (FICI) were calculated by using a checkerboard format. Biofilm formation was analyzed by using XTT-metabolic assay.
Results:
MIC of AmB for developing biofilm was lowered by 16 fold in combination with SNPs. The calculated fractional inhibitory concentration indices were 0.1875 suggesting that this interaction is synergistic. Similarly, the mature biofilms were significantly prevented by SNPs-AmB combination. This interaction was synergistic. Furthermore, interaction between SNPs and AmB against planktonic growth was additive. Hemolytic activity assay was carried out on these drugs and combinations. Drug required for inhibition alone as well as in combination did not exhibit hemolytic activity.
Conclusion:
The combinations with SNPs lead to decreases in the dosage of AmB required for anti-Candida activity. SNPs-AmB combination could be an effective strategy against biofilm formed by C. albicans.
1. INTRODUCTION
In the initial period of the 20th century, bacterial infections were a serious concern and a major cause of mortality. Oppositely, mycoses were not in attention but from the late 20th century mycoses are becoming a serious problem in immune-deficient patients such as AIDS, cancer, diabetes, cystic fibrosis, those who have undergone organ transplants and other invasive surgical operations [1, 2]. Fungal infections range from superficial, systemic infections to life-threatening invasive infections. When fungal cells invade the epithelial surfaces of immunocompromised hosts, it can lead to invasive fungal infections. These invasive fungal infections are associated with high morbidity and mortality. Pathogenic fungi causing invasive infections include Candida, Aspergillus and Cryptococcus [3, 4]. The major human opportunistic fungal pathogen, Candida albicans contribute to for approximately 50–60% cases of candidiasis particularly in immunocompromised patients. Azole drugs are commonly used to treat infections caused by C. albicans [5, 6].
Amphotericin B, (AmB) a drug developed in the 1950s is continued to be used against fungal infections in terminally ill patients [7, 8]. AmB kills yeast by binding to ergosterol in the fungal cell membrane and membrane permeabilization via channel formation [9, 10]. This drug is fungicidal in nature and can cause severe nephrotoxicity, chills and seizures in patients. In spite of the toxicity, AmB is still used in clinics because of its efficacy. There is a need for developing better drugs with less toxicity [11, 12]. Azole drugs even though effective are fungistatic in nature. Among the azole group, Fluconazole is the most widely prescribed drug [13]. Emergence of drug resistance among the major fungal pathogens of humans like C. albicans and formation of drug tolerant biofilms on abiotic as well as biotic surfaces in the human body is a serious current day concern. For example, C. albicans biofilms are resistant to most of the antifungal agents [14]. Silver nanoparticles (SNPs) are considered as an alternative by some workers [15].
In the present study, we have fabricated silver nanoparticles (SNPs) using the leaf extracts of a plant, Polyalthia longifolia. Synthesis of SNPs is standardized and the nanoparticles are characterized. We have explored the potential of using silver nanoparticles and AmB combinations against growth and biofilm forms of C. albicans. Purpose of this study is to bring down the MICs of both AmB and SNPs against growth, morphogenesis and drug resistant biofilm forms of C. albicans.
2. MATERIALS AND METHODS
2.1. Plant Material and Preparation of Extract
Mature fresh leaves of P. longifolia were collected from the campus of S.R.T.M. University, Nanded, Maharashtra, India. Taxonomic identification of the plant was done by Dr. R.M Mulani, Professor, School of Life sciences, SRTM University. The leaves of P. longifolia were thoroughly washed with distilled water and were blot dried with tissue paper. After drying the leaves were chopped into small pieces. Ten grams of chopped leaves were added in 100 ml of distilled water and boiled for five minutes to make an aqueous extract. The aqueous extract was filtered through a 0.45 μM membrane filter and used.
2.2. Synthesis of SNPs
Silver nitrate (AgNO3) was procured from Himedia Chemicals Ltd, Mumbai, India. Silver nitrate solution (1 mM) was prepared freshly for the synthesis of silver nanoparticles. The aqueous leaf extract of P. longifolia was used for the biogenic synthesis of the SNPs. One ml of the plant leaf extract was added to 19 ml of 1 mM aqueous silver nitrate solution in a 250 ml Erlenmeyer flask and incubated for 15 minutes at 70 °C. The biogenic synthesis of SNPs was observed spectrophotometrically at different time intervals. The biogenic synthesis of SNPs was observed by the color change from colorless to brown in the colloidal solution [16].
2.3. Characterization of SNPs
The synthesis of SNPs was characterized at different time intervals using a UV-Vis double beam spectrophotometer (Shimadzu UV-Visible Spectrophotometer) at the wavelength range 200-800 nm, and scanning interval was 0.5 nm. The surface morphology of biogenic synthesized SNPs was studied by high resolution scanning electron microscopy (SEM) (S-3700N: Hitachi, Japan). The structural analysis of functional groups present in the leaf extract of P. longifolia and SNPs biosynthesized from leaf extract of P. longifolia was done by Fourier transform infrared spectroscopy (FTIR) (8400 S; Shimadzu, Japan). Energy-dispersive X-ray (EDAX) analysis was done to determine the elemental composition (S-3400N; Hitachi, Tokyo, Japan). The crystal lattice and size of the biogenically synthesized SNPs were analyzed by X-ray diffraction (XRD) measurement using an XRD-6000 X-ray diffractometer (Shimadzu, Kyoto, Japan). The size of crystallite domain was calculated from width of obtained XRD peaks. The size distribution of SNPs was measured by dynamic light measurement (DLS) and zeta potential was measured using DLS (SZ-100; Horiba) [16].
2.4. Cultures, Culture Conditions, Media and Chemicals
C. albicans, ATCC 90028 was obtained from the Institute of Microbial Technology, Chandigarh, India. A clinical isolate of C. albicans GMC 03 was obtained from Government Medical College, Nanded, Maharashtra, India. The culture was maintained on Yeast extract-Peptone-Dextrose (YPD) (1% Yeast extract, 2% Peptone, and 2% Dextrose) agar slants at 4 °C. YPD was prepared and used for inoculum preparation. All the ingredients of Spider medium (1% Mannitol, 1% Nutrient broth, 0.2% K2HPO4), and 2, 3-bis (2-methoxy-4-nitro-sulfophenyl)-2H-tetrazolium- 5-carboxanilide (XTT) were obtained from Himedia Chemicals Ltd, Mumbai, India. Menadione was purchased from Sigma Aldrich Chemicals Ltd. India. A single colony from the YPD agar plates was inoculated in fifty ml of YPD broth and incubated at 30 °C on an orbital shaker, at 120 rpm for 24 h. Cells from the activated culture were harvested by centrifugation for 5 min at 2000 g, washed three times, and resuspended in PBS buffer (10 mM phosphate buffer, 2.7 mM potassium chloride and 137 mM sodium chloride, pH 7.4). Dimethyl sulphoxide was used as a solvent at a final concentration of 1%. Amphotericin B (AMPHOTRET) was purchased from Bharat Serums and Vaccines Limited, Mumbai, India.
2.5. Minimum Inhibitory Concentration for Planktonic Growth
The effect of SNPs on the growth of planktonic cells of C. albicans was studied by using the standard broth microdilution methodology based on the Clinical Laboratory Standards Institute guidelines [31]. Various concentrations of SNPs ranging from 0.75 to 100 µg ml -1 were prepared in Spider medium in non-treated 96 well polystyrene plates (Costar, USA). Wells without test compounds served as controls. 100 µl microlitres of inoculum were added to 100 μl of Spider medium in each well to obtain 1×103 cells ml-1. The plates were incubated at 35 °C for 48 hours. The growth was analyzed by taking the absorbance at 620 nm using a microplate reader (Multiskan EX, Thermo Electron Corp., USA). The lowest concentration of the SNPs which caused a 50% reduction in the absorbance compared to the control was considered as the minimum inhibitory concentration (MIC) for growth of C. albicans [17].
2.6. Minimum Fungicidal Concentration (MFC)
SNPs concentrations in the range 0.75 to 100 µg ml -1 were selected MFC testing. To determine MFC, Candida cells from the MIC and wells containing concentrations above the MIC were used. Aliquots of ten microlitres from these wells were spread on YPD agar. These plates were kept for incubation for 48 h at 30 °C and observed for growth. No appearance of colonies on the agar plates was noted as fungicidal effect [17]. The lowest concentration of the SNPs in the microtiter plate wells from which an aliquot showing no appearance of colonies was considered as the MFC.
2.7. Adhesion Assay
To investigate the effect of SNPs on the adherence of C. albicans to a solid surface (i.e. polystyrene) was studied using a microplate-based assay [17]. Various concentrations of SNPs ranging from 0.75 to 100 µg ml -1 were prepared in PBS. Wells without SNPs were maintained as controls. 50 µl of cell suspension was added to each well to obtain 1×107 cells ml-1. The final volume of the assay system in each well was kept hundred microlitres. The plates were incubated at 37 °C for 90 min at 100 rpm in an orbital shaking incubator to allow attachment of cells to the surface. After the incubation, wells were washed with PBS to remove nonattached cells. The density of the adherence in each well was analyzed as relative metabolic activity (RMA) using the XTT-assay. More than a 50% reduction in RMA compared to the control was considered significant.
2.8. Morphogenesis
The effect of SNPs on serum induced yeast to hyphal form morphogenesis was investigated in microtiter plate based assays [18]. Various concentrations of the SNPs were prepared in de-ionized distilled water with 20% serum. Cells of C. albicans were inoculated to obtain 1×10 6 cells ml -1 in the test and control wells. The final volume of the assay system in each well was maintained at 200 μl. The plates were incubated at 37 ° C on an orbital shaker at 200 rpm for 2 h. Cells were observed microscopically for the formation of germ tubes. The numbers of yeast cells and hyphae were counted out of 100 cells. The concentration which showed inhibition of hyphae by ≥ 50% compared to the control was considered the MIC for morphogenesis.
2.9. Biofilm Formation
C. albicans biofilms were developed on the surface of 96-well polystyrene plates as in a standardized in vitro biofilm model [19, 20]. Cell suspensions of 1×107 cells ml-1 were prepared in PBS and 100 µl were inoculated into each well. In the adhesion phase, plates were incubated at 37 °C for 90 min at 100 rpm to allow attachment of the cells to the solid surface. Non-adhered cells were removed by washing the wells with sterile PBS. Two hundred microlitres of the spider medium were then added to each well and the plates were incubated at 37 °C for 48 h to allow for biofilm formation. To analyze its effect on biofilm development, various concentrations of SNPs were prepared in spider medium and added to each well immediately after the adhesion phase and incubated for 48 h at 37 °C. To study the activity against mature C. albicans biofilms, SNPs were added to 24 h old mature biofilm and incubated for 48 h. After incubation, the wells were washed to remove any nonattached planktonic cells. Wells were observed for presence or absence of biofilms using an inverted light microscope (Metzer, India). Photographs were taken with a Labomed microphotography system (Labomed, India) at ×200 magnification. Biofilm growth was analyzed and confirmed with the XTT metabolic assay.
2.10. Biofilm Quantitation by XTT Assay
To quantify biofilm growth, XTT metabolic assay was performed [20]. The XTT solution was prepared by mixing 1 mg ml-1 of XTT salt in sterile distilled water and stored at -20 °C. Prior to use, menadione solution was prepared in acetone and added to the XTT to a final concentration of 4 μM. The wells containing biofilm were washed with PBS to remove non-adhered cells and incubated with 100 μl of XTT-menadione solution in dark, at 37 °C for 5 h. Color product by the water soluble formazan product was measured at 450 nm using a microplate reader (Multiskan EX, Thermo Electron Corp. USA). Wells without the SNPs were referred as controls. The concentration of SNPs which caused ≥ 50% lowering in relative metabolic activity was considered the MIC for biofilm formation.
2.11. Checkerboard Assay for Drug Combination Against Planktonic and Biofilm Growth
To study the efficacy of drug combination against planktonic and biofilm growth in C. albicans, checkerboard assay was performed and fractional inhibitory concentration indices (FICI) was analyzed [21]. A two dimensional array of drug combination was used for the preparation of drug dilutions. The efficacy of drug combinations against planktonic and biofilm growth was performed as discussed in the earlier section and the MIC values were determined. The FICI values were calculated by the formula:
ΣFIC = FICA + FICB. Where, FICA = (MIC of drug A in combination / MIC of drug A alone) FICB = (MIC of drug B in combination / MIC of drug B alone).
When the value of ΣFICI ≤ 0.5, it was considered as synergism; between 0.5 and 1.0, it was additive; and when ΣFICI >4, was considered as antagonism. A ΣFIC result of >1 but ≤ 4 was treated as indifference [22].
2.12. Scanning Electron Microscopy
Biofilms developed on oropharyngeal silicon disc were fixed in 2.5% glutaraldehyde (prepared in the PBS) at 4 ˚C temperature for 24 h and post-fixed in the 2% aqueous solution of osmium tetroxide for 4 h. The samples were dehydrated using a series of graded alcohols and finally dried to the critical drying point with a critical point dryer (CPD). The samples were mounted on the stubs and sputter coated with gold for 10 s with 11000 mA under the Argon plasma. A scanning electron microscope (Hitachi S- 3000N, Japan) operating under the high vacuum with 10kv was used to observe the biofilm structure [20].
2.13. Hemolytic Assay
The hemolytic activity of drugs was determined using mammalian red blood cells [23]. Human erythrocytes were collected. EDTA (1–2 mg/ml) was added in to the tube containing erythrocytes as an anti-coagulant. The erythrocytes were harvested by centrifugation for ten minutes at 634 × g at 20°C, and washed three times in saline. 10% (v/v) erythrocytes/saline suspension was prepared by adding saline to pellet. The 10% suspension was diluted 1:10 in saline in an Eppendrof tube. From different dilutions of SNPs and AmB prepared in saline, 100 µl of different concentrations of SNPs and AmB was added in triplicate to 100 μl of cell suspension. 1% Triton x -100 was used as a negative control and to achieve total hemolysis. Saline without any molecule was maintained as a positive control. The Eppendrof tubes were incubated for 1 h at 37°C and centrifuged for 10 min at 634 × g at 20°C. From the supernatant fluid, 150 μl was added to a flat-bottomed microtiter plate (Himedia Chemicals, India Ltd., India), and the absorbance was measured at 450 nm. Percent hemolysis was calculated using following equation:
Percentage hemolysis = [(Abs 414 nm in the compound solution – Abs 414 nm in PBS)/ (Abs 414 nm in 0.1% Triton X-100 – Abs 414 nm in Saline)] х 100
2.14. Statistical Analysis
Values presented are the means with standard deviations, obtained from three different observations. Values in the control and treatment groups for various molecules as well as the results obtained in XTT were compared using the Student’s t-test. A value of p < 0.05 was considered statistically significant [24] (Table 1).
| Sample | Plant Extract (in ml) | Silver Nitrate (in ml) |
|---|---|---|
| A1 | 0.25 | 19.75 |
| A2 | 0.5 | 19.5 |
| A3 | 1 | 19 |
| A4 | 1.5 | 18.5 |
| A5 | 2 | 18 |
| A6 | 3 | 17 |
| A7 | 4 | 16 |
| A7 | 5 | 15 |
3. RESULTS
3.1. Characterization of SNPs
Biogenic synthesis of SNPs was done using different concentrations of the leaf extract of P. longifolia. A change in color was observed in the reaction mixture from colorless to pale yellow which later changed to brown color. The aqueous silver nitrate solution without plant extract did not show any change in color hence served as blank reference. The presence of SNPs was monitored by UV-Vis spectrometer analysis. As the concentration of the plant extract increased up to 1ml, there was an increase in the absorbance (Additional File 1; Figs. 1A, B and C). Maximum enhancement of synthesis of SNPs was observed for the reaction of 1 ml of leaf extract in a total reaction mixture of 20 ml with incubation time of 24 hrs. As substrate concentration i.e. silver nitrate increased up to 3 mM, there was an increase in absorption of SNPs (Additional File 1; Figs. 1E, F and G). However, reduced absorption of SNPs was observed at 4 and 5 mM of substrate concentration and peak shifted towards 450 nm. Absorption spectra of SNPs synthesized using 1mM and 2 mM showed absorbance 2.78 at 432 nm and 3.01 at 435 nm respectively. As we increased the temperature of reaction mixture up to 70 0 C, the rate of the synthesis of SNPs was boosted (Additional File 1; Figs. 1H, I, J, K, and L).
| Sr. No. |
Wavelength of X –ray source (λ in nm) |
Scherer Constant (K) | Full width at Half Maximum of Diffraction Peak (β) | 2 θ (in deg) | FWHM (in deg) | Crystallite Size of SNPs (in nm) |
|---|---|---|---|---|---|---|
| 1 | 0.1541 | 0.9 | 0.00214 | 38.067 | 0.2495 | 68.70 |
| 2 | 0.1541 | 0.9 | 0.00261 | 44.23 | 0.2999 | 57.4 |
| 3 | 0.1541 | 0.9 | 0.003951 | 64.437 | 0.4529 | 41.5 |
| 4 | 0.1541 | 0.9 | 0.00517 | 77.374 | 0.5923 | 34.4 |
FTIR spectrum of biogenically synthesized SNPs and plant extract showed peaks at 3433.5 (O-H stretching in phenol or carboxylic group), 2924.5 (alkyl C-H stretch), 2854.6 (alkane C-H stretch), 1629.7 (amide C=O stretch), 1448.29 (–C–C– stretch (in ring) aromatic), 1020.94 cm-1 (C-O-C stretching aromatic ring) (Figs. 02A and B). We compared XRD spectrum of SNPs with standard which revealed that SNPs formed biogenically were crystalline in nature. We noticed peaks at 2θ values of 38.0670, 44.230, 64.430 and 77.37 0 corresponding to (111), (200), (220) and (311) Bragg reflections, respectively (Fig. 03A). Unsigned peaks for silver were observed in spectra denoted by (*) in (Fig. 3A) which may be related to crystalline and amorphous organic phases. The average particle size of SNPs was calculated by using Debye-Scherrer equation. The calculated average particle size of SNPs was 50.5 nm (Table 2). SEM analysis of bio synthesized SNPs showed widely distributed silver nanoparticles were circular. The Nanoparticles formed were in different sizes ranging from 50 -70 nm (Fig. 3B). DLS and zeta potential analysis of biosynthesized SNPs revealed that SNPs are having average size of 29.3 nm and the nanoparticles possess charge of -23 mV (Figs. 4A, B). The EDAX analysis showed the presence of strong elementary silver signal of SNPs that confirms the presence silver. EDAX characterization also showed weak signals for other atoms like oxygen which may derived from biomolecules used for synthesis and capping of SNPs from plant extract (Fig. 5).
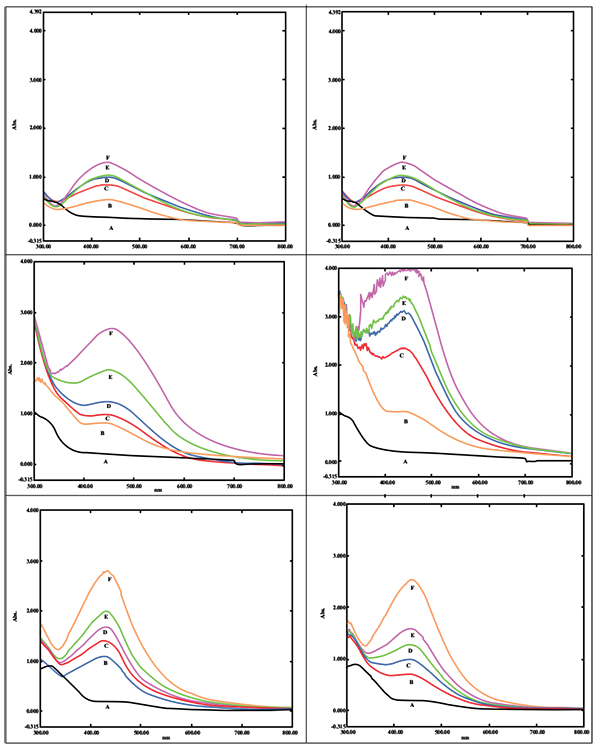 |
Additional File 1: UV-visible spectrum of SNPs. |
 |
Fig. (2). Fourier transform infrared spectroscopy spectrum of plant extract (A) and biosnthesized SNPs (B). |
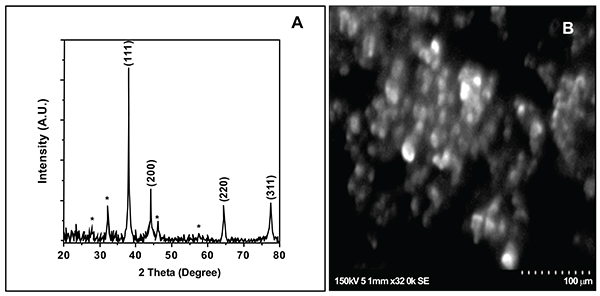 |
Fig. (3). X-ray diffraction spectrum (A) and scanning electron microscopy analysis (B) of biosynthesized SNPs. |
3.1.1. Antifungal Activity of SNPs Against Growth and Various Virulence Factors of C. Albicans
SNPs caused inhibition of planktonic growth of C. albicans in a concentration-dependant manner. At 1.5 µg/ml and 3 µg/ml, SNPs caused significant reduction in planktonic growth of C. albicans ATCC 90028 and GMC 03 respectively (Fig. 6). SNPs at 50 and 25 µg/ml exhibited fungicidal activity against C. albicans ATCC 90028 and GMC 03 respectively (Table 3). SNPs inhibited the yeast to hyphae morphogenesis in C. albicans (Fig. 6). C. albicans cells treated with 1.5 and 3.13 µg/ml of SNPs totally disrupted hyphal transition in C. albicans ATCC 90028 and GMC 03 respectively (Fig. 7). C. albicans cells exposed to 6.25 and 3.13 µg/ml concentrations of SNPs significantly inhibited adhesion of cells on polystyrene surfaces of C. albicans ATCC 90028 and GMC 03 respectively (Fig. 6). C. albicans treated with of 50 µg/ml of SNPs showed prominent inhibition in early biofilm development. At this concentration percentage, metabolic activity of biofilms was reduced by 80-85% (Fig. 6). The concentration required for the inhibition of mature biofilm formation was higher as compared to concentration required for inhibition of early biofilm formation. At 100 µg/ml concentration of SNPs, mature biofilm formation was significantly reduced in both the strains of C. albicans (Fig. 6). Control biofilms were observed to be multilayered, characterized by a dense network of hyphae. Biofilms treated with SNPs exhibited reduced cell density, defective filaments and consisted of a layer of yeast cells (Fig. 8).
3.1.2. Efficacy of SNPs-AmB Combination Against Planktonic and Biofilm Forms of Growth
MIC of SNPs for the inhibition of planktonic growth of C. albicans ATCC 90028 was 1.5 µg/ml. SNPs at a concentration 0.187 µg/ml when combined with 0.125 µg/ml of AmB inhibited the growth of C. albicans significantly. Addition of SNPs caused 2 fold decrease in the MIC of AmB i.e. MIC decreased from 0.25 µg/ml to 0.125 µg/ml (Table 4). FICI index was 0.6246 suggesting that interaction was additive. Similarly, the combination of SNPs and AmB showed pronounced effect against C. albicans GMC 03. MIC of AmB was reduced in the presence of SNPs by 15 fold i.e. 0.25 to 0.016 µg/ml. The combination of 0.375 µg/ml of SNPs with 0.016 µg/ml of AmB showed pronounced effect against planktonic growth of GMC 03. The FIC index for combination was 0.189 indicating synergistic interaction (Table 4).
MIC of AmB was achieved at 1 μg/ml against the developing biofilm of C. albicans. SNPs showed significant effect against developing biofilm at 50 µg/ml. SNPs at a concentration 3.125 μg/ml and 0.125 μg/ml of AmB combination prevented the development of early biofilm in C. albicans ATCC 90028. MIC of AmB was decreased by 16 times on the addition of SNPs i.e. 1 to 0.125 µg/ml. FICI was 0.1875 inferring that interaction is synergistic (Table 4). In the clinical isolate of C. albicans GMC 03, FICI for interaction between SNPs and AmB was 0.75 indicating that interaction was additive (Table 4).
MIC of SNPs for mature biofilm formation was achieved at 100 µg/ml whereas the MIC of AmB for mature biofilm formation was 2 µg/ml. MIC of AmB was lowered by four fold i.e. 2 to 0.5 µg/ml in the presence of SNPs for mature biofilm in C. albicans ATCC 90028. The FICI was 0.3125 suggesting that interaction was synergistic. In C. albicans GMC 03, SNPs-AmB interaction against mature biofilm was additive. The calculated FICI was 0.5625 (Table 4).
3.2. Toxicity Studies of SNPs and AmB Using Mammalian Erythrocytes
Hemolytic activity of SNPs and AmB was evaluated using human erythrocytes. SNPs concentrations ranging from 0.78 to 25 µg/ml did not cause significant hemolysis and >94% of human erythrocytes did not show hemolysis. SNPs in the concentration range of 25 and 50 µg/ml caused significant hemolysis (Fig. 9A and B). AmB at 8 µg/ml exhibited 11% of hemolysis whereas AmB in the concentration range from 0.0625 to 4 µg/ml caused negligible hemolysis (in the range of 1-5%) (Fig. 9B).
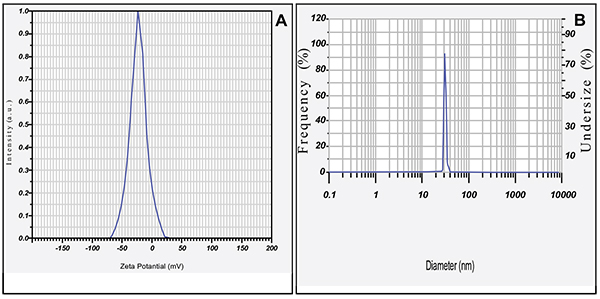 |
Fig. (4). Zeta potential analysis (A); dynamic light scattring graph (B) of biosynthesized SNPs. |
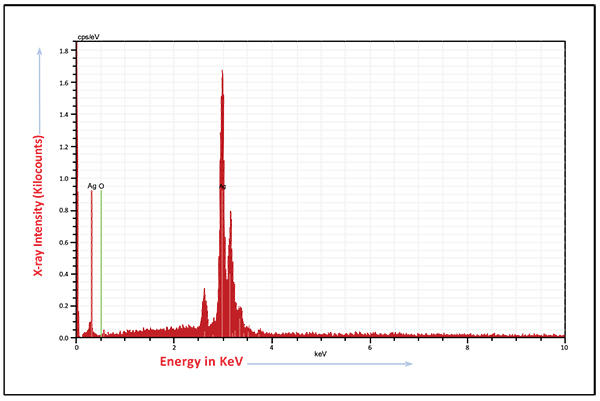 |
Fig. (5). Energy-dispersive X-ray spectrum of biosynthesized SNPs. |
4. DISCUSSION
The use of biofabricated SNPs in various technologies is becoming important due to the ecofriendly way used for its synthesis and as such there is great demand to develop green technology for the synthesis of nanomaterials [25]. P. longifolia is well-known for its medicinal properties and is also reported for its antibacterial and antifungal properties [26]. However, there are no reports available on the anti-Candida activities of SNPs synthesized using this plant. Kavivya et al. (2011) reported the antibacterial activity of SNPs synthesized from this plant [27]. We synthesized SNPs using the leaf extract of P. longifolia, which is a one step, non-toxic and environmentally friendly way for production. The exact mechanism behind biosynthesis of silver nanoparticles is not known. But it is reported that phytochemicals in the plants may be responsible for the reduction of silver ions to silver nanoparticles [28].
The mechanism behind the antimicrobial action of nanomaterials seems to be complex and may involve several mechanisms including the production of reactive oxygen species (ROS), cell membrane disruption, mitochondrial as well as DNA damage [29-32].
In the present study, we have explored the efficacy of biosynthesized silver nanoparticles against growth and different virulence factors of C. albicans. Yeast to hyphal form transition in C. albicans is a crucial factor in virulence, tissue invasion and biofilm formation [33]. We achieved the inhibition of yeast to hyphal morphogenesis on treatment with biosynthesized silver nanoparticles at 3 µg/ml (C. albicans ATCC 90028) (Table 3). This anti-morphogenetic activity of silver nanoparticles may lead to the prevention of the formation of biofilm. C. albicans is significantly involved in the biofilm formation on medical devices such as central venous catheters, joint prostheses. Infections caused by biofilms are difficult to treat as these are naturally resistant to antifungal agents such as Fluconazole [34, 35]. AmB inhibited both planktonic growth and biofilm formation (early and mature). AmB inhibited planktonic growth at 0.25 µg/ml in two strains of C. albicans. Prevention of early and mature biofilms required 1 and 2 µg/ml of AmB in both the strains respectively (Table 4). The biosynthesized silver nanoparticles showed significant inhibitory activity at 50 µg/ml and 100 µg/ml against early and mature biofilm in ATCC 90028 and GMC 03 respectively (Fig. 6). In addition to this, biosynthesized nanoparticles were fungicidal in nature (Table 3). Monteria and his coworkers (2011) have shown the efficacy of chemically synthesized silver nanoparticles having an average size of 5 nm against the adhered cells and biofilm of C. albicans [36]. From various studies exhibiting contrasting results it may be said that the anti-Candida properties of silver nanoparticles may depend on the mode of synthesis, size, shape, stabilizing agent, application of method etc [37]. The effect of commercial silver nanoparticles functionalized with polypropylene is reported by Vazquez-Munoz (2014). Silver nanoparticles accumulated outside the cells and released silver ions into the cells. The fungicidal activity seems to be due to release of silver ions from the silver nanoparticles [38].
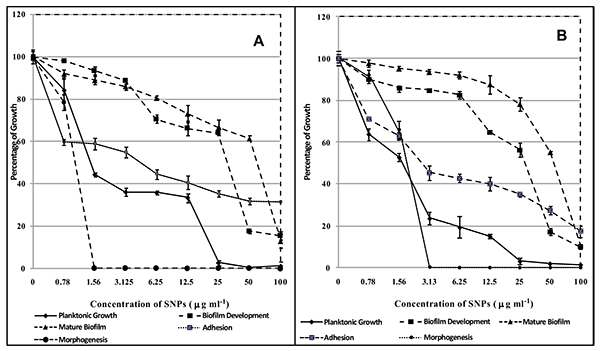 |
Fig. (6). Effect of biosynthesized SNPs on planktonic growth adhesion, morphagenesis, developing biofilm and mature biofilm of (A) Candida albicans ATCC 90028 (B) Candida albicans GMC 03. |
| MIC (µg/ml) | MFC (µg/ml) | |||||
|---|---|---|---|---|---|---|
| Candida albicans | Planktonic Growth | Morphogenesis | Adhesion | Biofilm Development | Mature Biofilm | |
| ATCC 90028 | 1.56 | 1.56 | 6.25 | 50 | 100 | 25 |
| GMC 03 | 3.13 | 3.13 | 3.13 | 50 | 100 | 50 |
Many in vitro studies of antifungals agents have revealed that synergistic combinations can give better antifungal activity, reduces the risk of drug resistance and lowered toxicities [39]. Hari et al. (2014) carried out synergistic interaction studies on antibacterial agents and silver nanoparticles against Escherichia coli, Bacillus subtilis, Lactococcus lactis, Pseudomonas aeruginosa, Micrococcus luteus, Staphylococcus aureus, and Proteus vulgaris. It was proposed that, the synergistic interaction may due to binding of silver nanoparticles to the antibiotics that may attach to cell membrane and result in membrane damage. Further silver nanoparticles may combine with antibiotics and enter the cells and finally result in cell death by prevention of DNA unwinding [40]. In comparison with antibacterial study, antifungal studies in combination with the silver nanoparticles are limited. However, the combined effect of Fluconazole with mycofabricated silver nanoparticles on the growth and biofilm formation of C. albicans is reported. These studies suggested that, combination of silver nanoparticles results into enhanced effect of Fluconazole against growth of C. albicans [41]. Nystatin combination with silver nanoparticles was found to act synergistically against biofilms of C. albicans and C. glabrata [42]. Higher antifungal activity of AmB silver nanoparticles hybrid is reported against Aspergillus niger, C. albicans and Fusarium culmorum [43]. AmB is widely studied in combination with other antifungal agents other fungicidal agents [44]. Recently, Pd@Ag nanosheets showed synergistic anti-cryptococcal fungicidal effects in combination with AmB [45]. Saldanha et al. (2016) reported nanocomplex comprised of AmB and nanosized magnetite exhibited antifungal activity against Paracoccidioides brasiliensis with low cytotoxicity [46]. Tyrocidies exhibited synergistic activation in combination with AmB in biofilm eradicating activity in C. albicans [47]. Sophorolipid when combined with AmB acts synergistically against biofilm formation in C. albicans [48]. A new form AmB-Cu2+ ions complex is reported to exhibit less toxic effects on human renal cell by changing the expression of tumor necrosis factor (TNF) receptors and genes involved in TNF induced pathways [49]. Synergistic combination of Nitroimidazoles C- AmB, Histatin 5 -AmB, Curcumin -AmB, AmB- Terbinafine, Farnesol -AmB, Eugenol C AmB,, Baicalein-AmB, Anidulafungin -AmB, Aspirin-AmB, AmB- Drospirenone, AmB-N-acetylcysteine, AmB-Rifampicin, AmB-Caspofungin, Berberine -AmB, Cilofungin -AmB, AmB- Ketoconazole, AmB-Azoles and Mycograb-lipid-associated AmB has been reported against C. albicans [50].
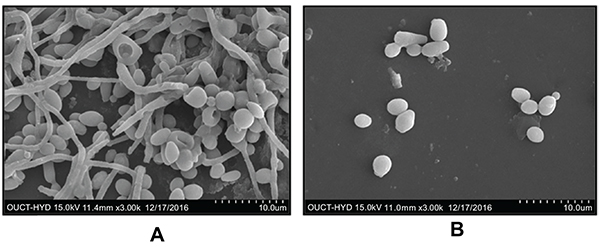 |
Fig. (8). Scanning electron micrographs of silver nanoparticles (SNPs) against biofilm development of C. albicans (ATCC 90028) (A) control; (B) 50 µg/ml. |
Here, for the first time we are reporting synergistic and additive interaction of biosynthesized silver nanoparticles with AmB against planktonic and biofilm forms of C. albicans. Inhibition of planktonic growth of C. albicans required 0.187 µg/ml of SNPs with 0.125 µg/ml of AmB. Two fold decrease in MIC of AmB was observed in presence of SNPs. Interestingly, the calculated FICI was 0.624 indicating that interaction was additive. However, this interaction was synergistic in GMC 03 suggesting that there is strain variation in response of AmB and SNPs combination. The combination study on SNPs-AmB against developing and mature biofilm reveals that the interaction was synergistic and additive. In the combination study carried out for SNPs –AmB against developing biofilm, we found that, MIC of AmB was decreased by 16 fold and the interaction was synergistic. Similarly, SNPs-AmB combination was synergistic against mature biofilm. However, this interaction was additive in the clinical isolate GMC 03 (Table 4). We can infer that, there is strain variation in response of SNPs-AmB combination.
| Growth | MIC (µg/ml) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Candida albicans ATCC 90028 | Candida albicans GMC 03 | ||||||||||||
| Alone | In Combination | Alone | In Combination | ||||||||||
| SNPs | AmB | SNPs | AmB | FICI | Remarks | SNPs | AmB | SNPs | AmB | FICI | Remarks | ||
|
Planktonic Growth |
1.5 | 0.25 | 0.187 | 0.125 | 0.624 | Additive | 3 | 0.25 | 0.375 | 0.016 | 0.189 | Synergistic | |
|
Biofilm Formation |
50 | 1.00 | 3.125 | 0.125 | 0.187 | Synergistic | 50 | 1.00 | 25 | 0.25 | 0.75 | Additive | |
|
Mature Biofilm |
100 | 2.00 | 6.25 | 0.5 | 0.312 | Synergistic | 100 | 2 | 6.25 | 1.00 | 0.562 | Additive | |
Available studies suggest that the antimicrobial activity of silver nanoparticles may be due to changes in cell permeability, respiration, generation of free radicals, reaction with sulphur containing proteins, prevention of DNA from unwinding and finally cell death [51-55]. The mechanism behind anti-Candida activity may be similar. AmB acts on fungal cell membrane ergosterol. It binds to ergosterol and forms aggregates in transmembrane channels that may lead to leakage of cytoplasmic content and finally cell death [56-59]. The synergistic action of AmB-SNPs seems to involve several mechanisms and may be complex.
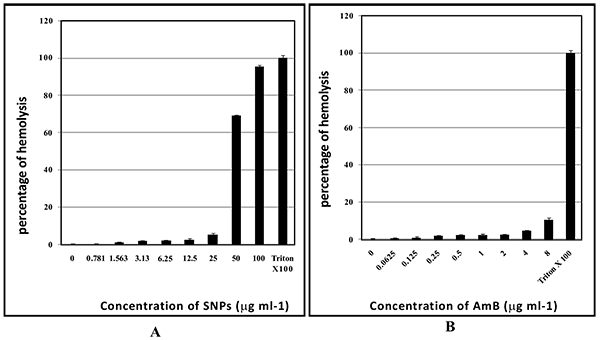 |
Fig. (9). Hemolytic activity of SNPs (A); and AmB (B). |
Silver nanoparticles are well known for its toxicity effects such genotoxicity and mitochondrial dysfunction [60, 61]. AmB at high concentrations may cause side effects such as nausea, vomiting, rigors, fever, hypertension or hypotension, and hypoxia. AmB can also lead to the side effects such as renal insufficiency, hypokalemia, hypomagnesaemia, metabolic academia, and polyuria due to nephrogenic diabetes insipidus [12]. Factors such as male gender, higher average daily dose of AmB (≥ 35 mg/day), diuretic doses, bodyweight (≥ 90 kg), other nephrotoxic drugs and abnormal renal function increase risk of AmB nephrotoxicity [58, 60, 61]. The ability to cause hemolysis can limit the therapeutic application of a drug. SNPs caused significant hemolysis at 50 and 100 µg/ml. Unfortunately, these concentrations are required for the prevention biofilms. As such SNPs application to clinical practices is not recommended. Combination with AmB with biofabricated SNPs leads to lowering down of the MIC of SNPs by 16 fold (Table 4). Interestingly, these concentrations are not exhibiting hemolysis at this concentration. So it may be safer for use.
CONCLUSION
We have systematically evaluated the potential of biosynthesized SNPs against growth and virulence factors of C. albicans such as morphogenesis, adhesion and biofilm formation. The SNPs –AmB combination showed pronounced effects on growth and biofilm formation. Main thrust of this work is lowering the dosages of AmB as well as SNPs required for anti-candida activity so that one can reduce the side effects. Hemolytic activity carried out on these drugs show that the combination does not exhibit hemolytic activity. The use of SNPs-AmB combinations seems to be a promising strategy for the inhibition of drug resistant biofilm forms of C. albicans. However, further, in vivo and toxicity studies are required.
AUTHORS CONTRIBUTION
SDH was responsible for all experimental work, data acquisition and analysis and writing and preparation of manuscript. SPM was contributed to experiments and data analysis. SMK was responsible for study concept, designing and coordinating the research, supervising the work and revising the manuscript. All authors read and approved the final manuscript.
LIST OF ABBREVIATIONS
| AmB | = Amphotericin B |
| SNPs | = Silver Nanoparticles |
| SEM | = Scanning Electron Microscopy |
| FTIR | = Fourier Transform Infrared Spectrometry |
| EDAX | = Energy-dispersive X-ray |
| XRD | = X-ray Diffraction |
| DLS | = Dynamic Light Measurement |
| MIC | = Minimum Inhibitory Concentration |
| MFC | = Minimum Fungicidal Concentration |
| ATCC | = American Type Culture Collection |
| FICI | = Fractional Inhibitory Concentration Index |
| XTT | = 2, 3-Bis (2-methoxy-4-nitro-5-sulfo-phenyl)2-Htetrazolium- 5-carboxanilide |
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No Animals/Humans were used for studies that are base of this research.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
SDH is thankful to UGC-Innovative program, New Delhi for providing financial assistance.






